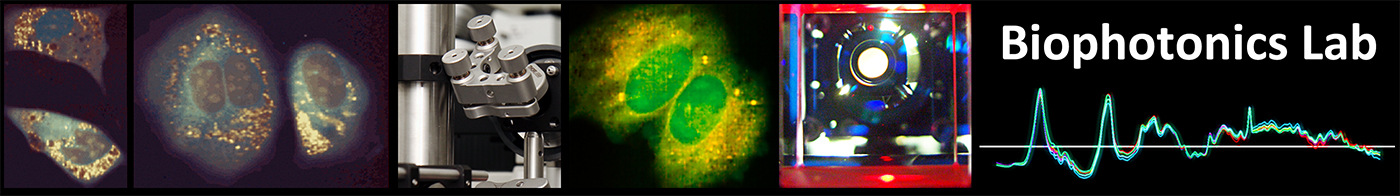
Compressed sensing
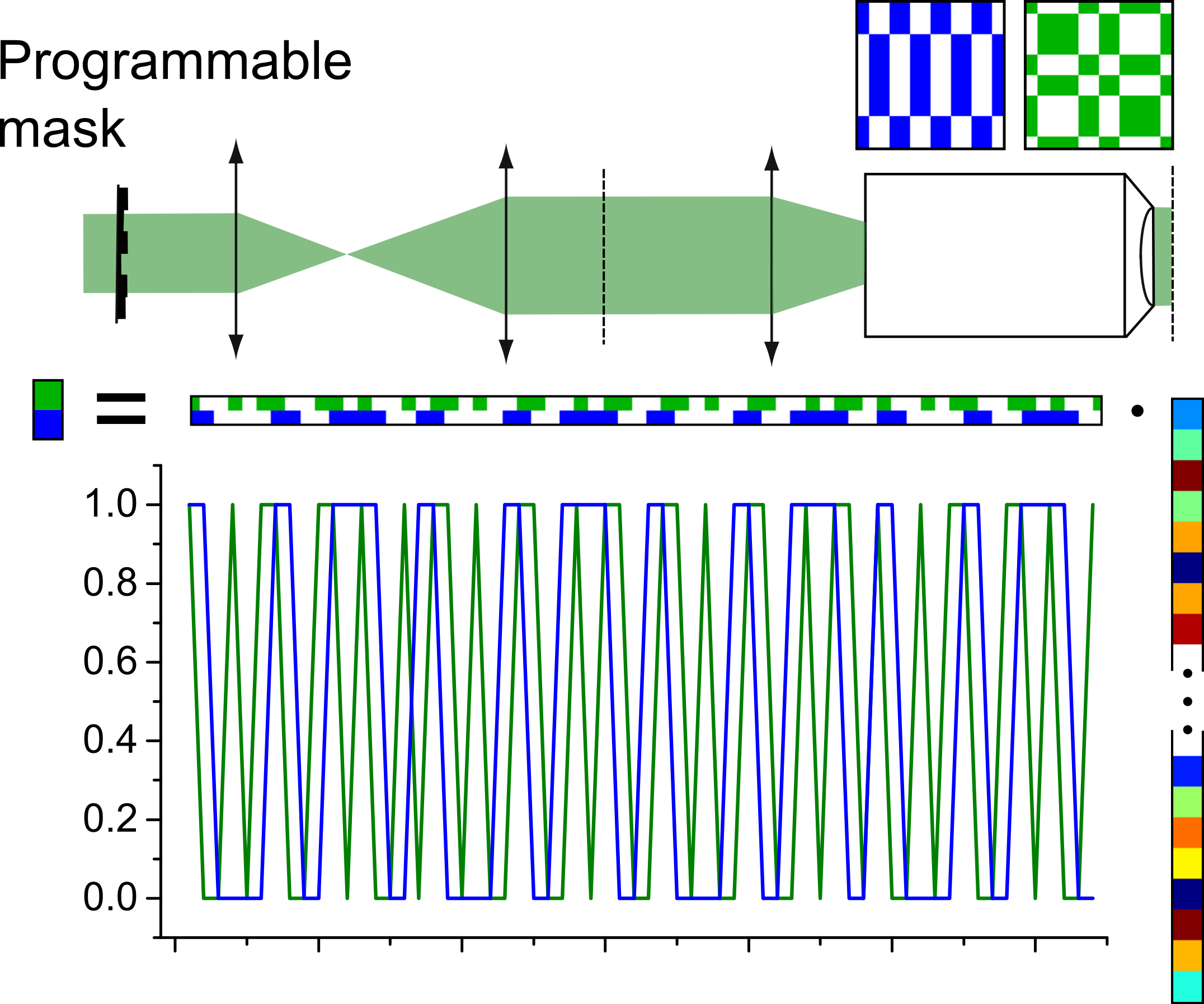
Standard CS based on the projection of known patterns.
Compressed sensing is a data acquisition approach that enables the recovery of a signal based on a significantly smaller amount of coefficients than the original amount of data points. The data collection strategy relies on projecting patterns onto the object and recording the resulting values. In the case the projected patterns form a suitable basis representing the underlying signal, reconstruction is possible based on certain assumptions of continuity of the signal.
While this approach enables very effective data compression compared to standard sampling, it possesses some limitations for microscopy techniques:
- It requires a dedicated system to enable the modulation of the illumination light, making it not usable with standard microscopes.
- It is inherently based on wide-field illumination, so that it is not applicable for advanced methods such as confocal or two-photon microscopy.
Compressed sensing applied to laser-scanning microscopes by including a model of the PSF.
We developed an approach where compressed sensing can be used for point-scanning system, by also including a model of the imaging system into the reconstruction algorithm. The point-based detection that is used in laser-scanning microscopy is not compatible with compressed sensing, as a linear combination between different image points based on the pattern basis is required for reconstruction. However, when considering the point-spread function (PSF) of the microscope, each point measurement effectively spreads over several pixels in the image, which creates a linear combination which relies purely on the physical properties of the microscope.
This approach makes it possible to employ compressed sensing even with commercial laser-scanning systems, where the sub-sampling is directly performed in the spatial domain, which directly leads to faster acquistion as well as reduced photo-bleaching. High-resolution images can then be reconstructed based on a total-variation minimization algorithm while preserving the spatial resolution.
 |
 |
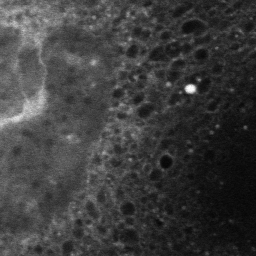 |
| Mouse embryonic fibroblasts cells tagged with an endoplasmic reticulum tracker, measured at low resolution (left, 256x256), that can be reconstructed with finer sampling (1024x1024) without loss of spatial resolution (middle) compared to a high-resolution image taken on the same cell (right). | ||
Code distribution
The code is based on the TVAL3 solver, initially developed at Rice University in the CAAM department. As the original code, this package is distributed under the GPL license (see LICENSE and README files in the package). The current version of the package is 1.1.
The package also provides example codes which relies on example data which is distributed separately, for both confocal microscopy and Raman micro-spectroscopy.